CRC Atlas is an interactive web server that is built on R Shiny for researchers to investigate landscape of gene-coexpressing modules (GEM) in curated single cell RNAseq (scRNAseq) data of colorectal cancer. In CRC Atlas, we have collected 279 samples and 626858 cells from 8 scRNAseq datasets. Please refer to citation for detailed information. (biorxiv link if applicable)
CRC Atlas | © DBMI 2023 | University of Pittsburgh
 Contact: Han Zhang (haz96@pitt.edu), Dr. Lujia Chen (luc17@pitt.edu)
Contact: Han Zhang (haz96@pitt.edu), Dr. Lujia Chen (luc17@pitt.edu)
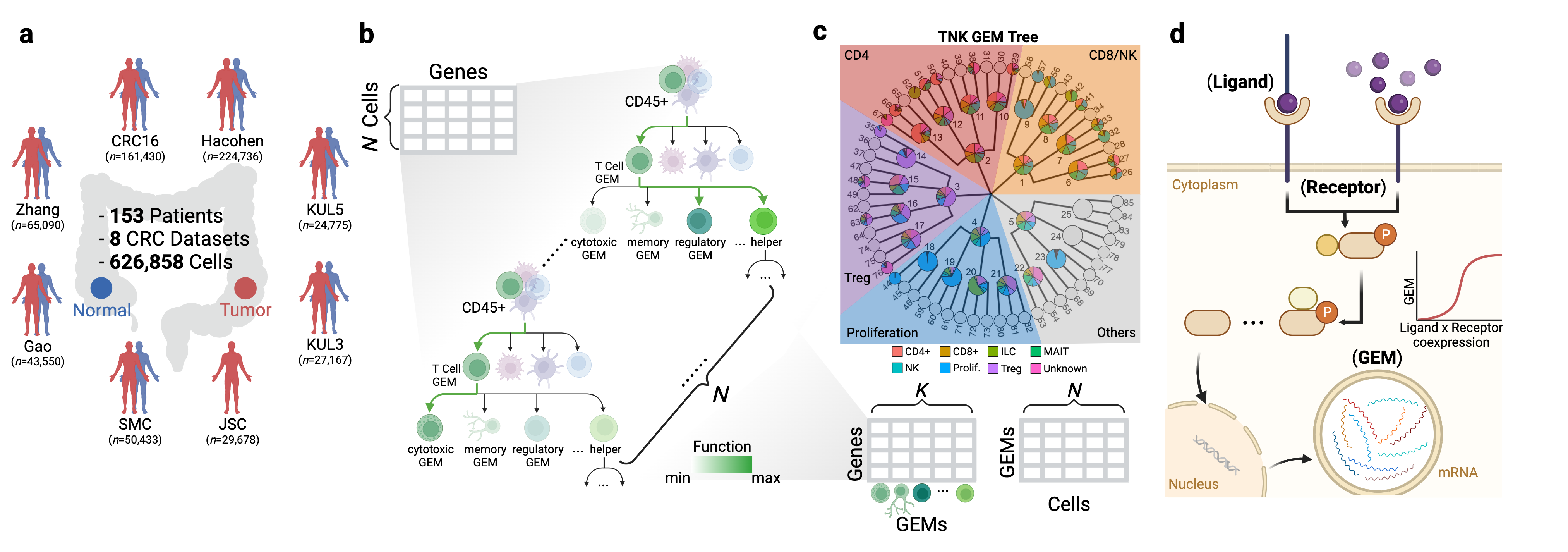
Look into the distribution of each gene-coexpressing modules (GEM) and associate top weighted genes by selecting GEM in below dropdown menu. Left panel shows the UMAP, top 20 genes and distribution of the inquired GEM over cell subtypes. Right panel displays the dot plots of all useful GEMs of this major cell type.
Look into the nonlinear Sigmoid relationship between gene-coexpressing modules (GEM) and ligand -> receptor (LR) pair in our curated CRC Atlas (n=151). On the right panel, you can also find the Top 10 significant GEM-LR pairs for your search, ranked by RSE (residual standard error).
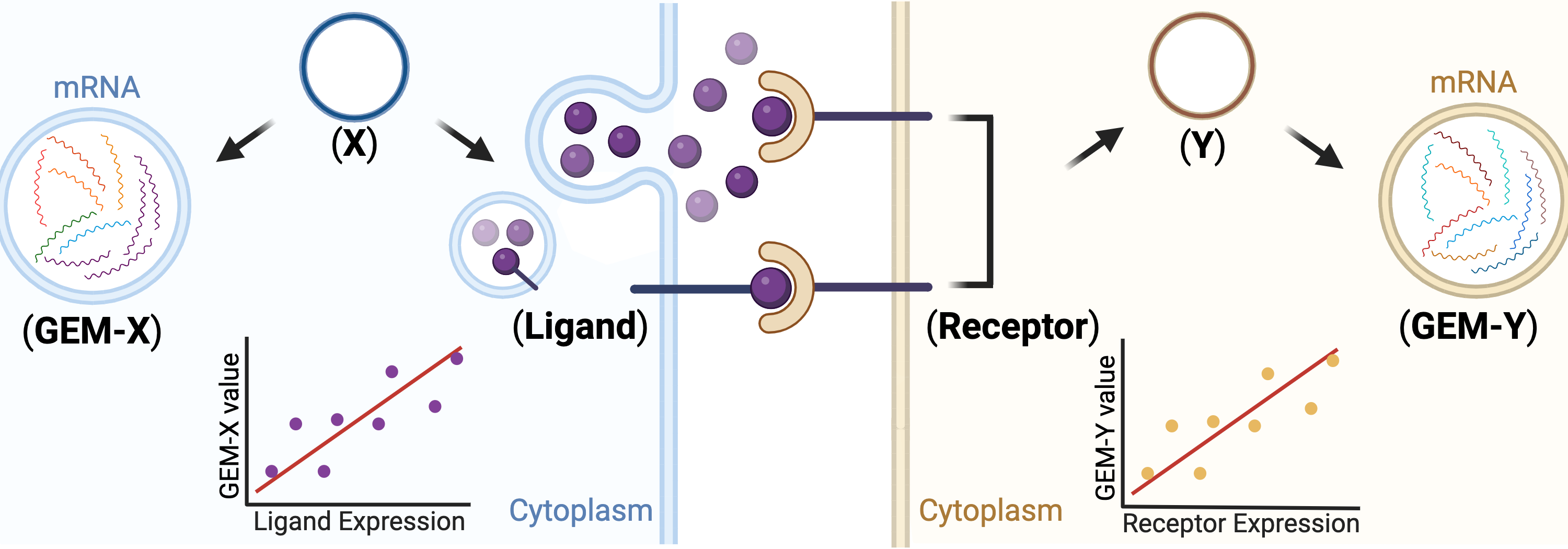
Conditional independence is an important concept in causal analysis, and in our setting, if a pair of highly correlated GEMs becomes independent when conditioning on a ligand -> receptor (LR) pair, the LR is likely involved in transmitting signal between cells expressing the GEMs. Finding a GEM X-LR-GEM Y triplet leads to the hypothesis depicted in below figure. Herein, we provide the triplet results that pass the conditional independence test. (choose at least one)
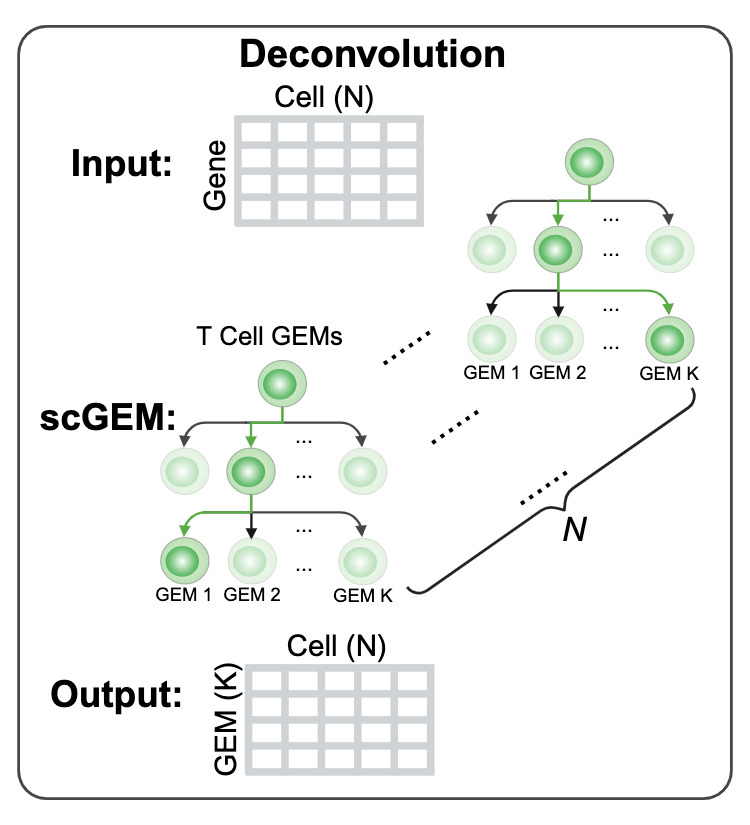
To deconvolute the bulk-RNAseq expression data using our curated GEM gene list, you will need to upload the gene expression in <.csv> format of which the rowname is gene symbol and colname is sample ID. The data is better deconvoluted if the input matrix is log2(TPM+1) or log2(FKPM+1).
Click here to download an example file